projects
Exploring and engineering resilient genomes for advanced animal and plant engineering
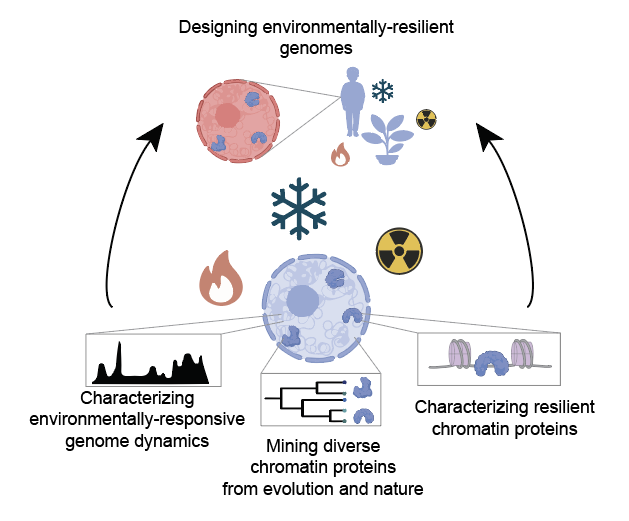
Diverse organisms are capable of surviving at extreme temperatures; however, the mechanisms that underlie the response and resiliency of the genome to these harsh conditions are poorly understood. A complete understanding of the molecular components of these mechanisms and how they have evolved to achieve their specialized functions would allow us to engineer chromatin to operate in extreme environments. I will leverage my expertise at the interface of evolution, synthetic biology, and environmental physiology to define the molecular underpinnings of this “genomic resilience”. Using a platform for the systematic, high-throughput functional, spatial, and genomic characterization of chromatin-associated proteins developed in my postdoctoral work, I will systematically define the design rules for resilient genomes, map how these rules have evolved and diversified over time, and use these insights, combined with computational protein design, to create novel protein components that can be used to bolster resilience in human and plant cells. This work will drive forward our understanding of molecular evolution, unveil fundamental physical mechanisms underlying genome organization, and inform the design and engineering of genomes for a broad range of cellular applications, including cryotherapies, tissue preservation, and environmentally resilient plants.
Postdoctoral work
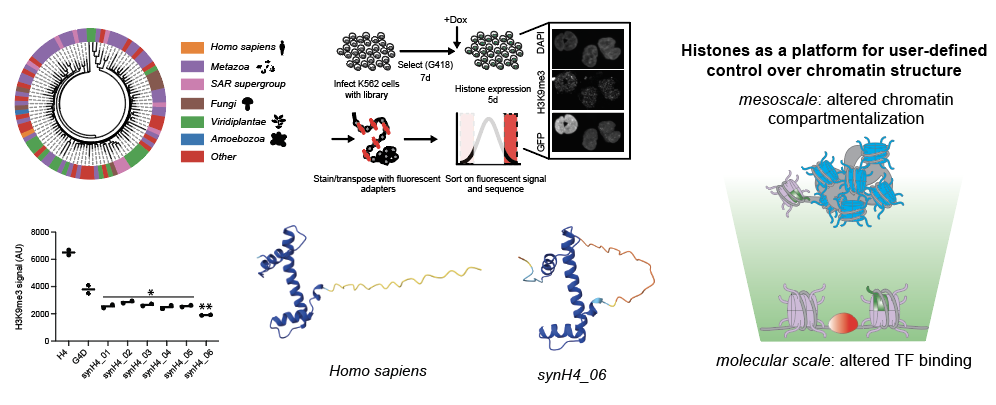
In my postdoctoral work, I explored how mesoscale chromatin organization can be encoded in histone proteins, which form nucleosomes, the building blocks of chromatin. Using deep mutational scanning, microscopy, and genomics, I discovered that even single amino acid changes in histones can dramatically reshape nuclear organization and transcription. I trained computational models to predict these effects from data, and used them to design synthetic histones that drive targeted epigenetic states (Jena et. al. 2025). This work positions chromatin not just as a structural framework or readout of cell identity, but as an engineerable scaffold for driving new cellular functions.
Previous work
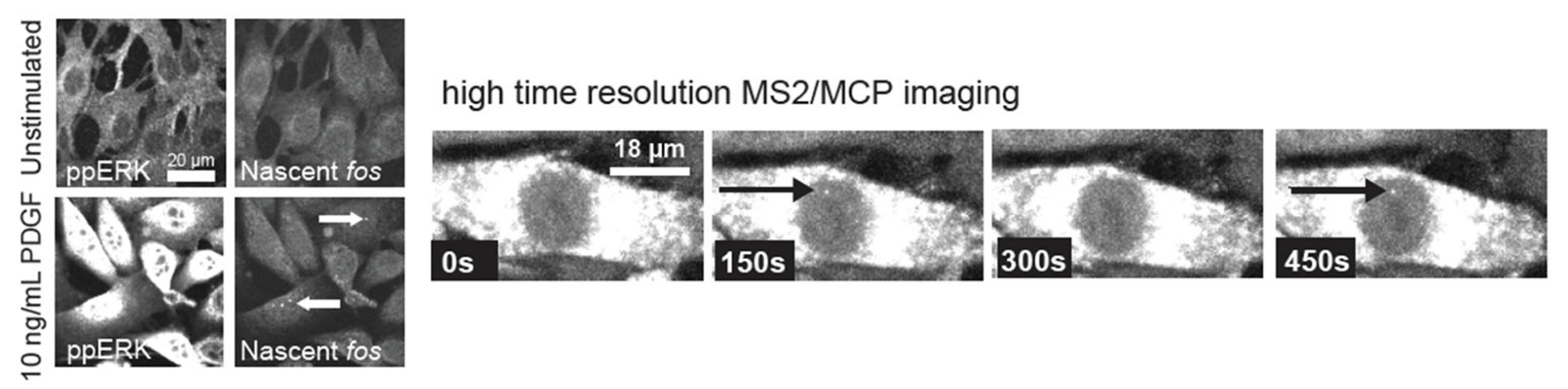
In my PhD, I primarily focused on the recently discovered phenomenon of Erk signaling dynamics, where the Erk pathway activates and deactivates in a pulsatile fashion. I asked two main questions in my work: 1. How does such rapid pulsatile behavior feed forward to the level of Erk-responsive gene activation? and 2. How can we infer the sources of signaling pulses in a population of cells that display both cell-autonomous and cell-to-cell coupling behavior in the same pathway? My work uncovered a set of amplitude, duration, and context-dependent rules for fractional transcriptional responses to transient Erk signaling (Jena et. al. 2021). In addition, my collaborators and I established a set of models and accompanying machine learning tools for inferring signaling behavior from live-cell imaging (Verma*, Jena* et. al. 2021).
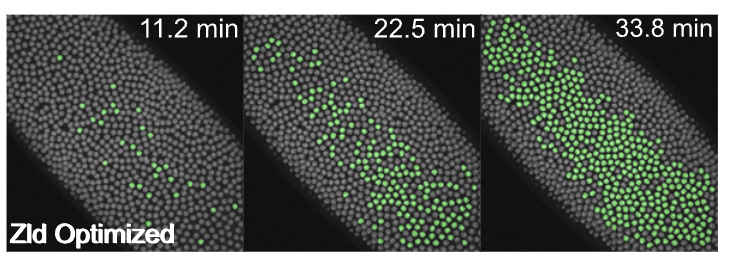
My work connecting time-dependent signaling to gene regulation has also extended to developmental, whole-animal contexts, where I dissected the contributing enhancer elements that allow for rapid interpretation of an upstream transcription factor gradient in the fly embryo (Keller*, Jena* et. al. 2020).
My undergrad thesis, completed under the mentorship of Martin Karplus, was focused on using molecular dynamics to simulate the effect of membrane lipid composition on the behavior of embedded proteins. I also studied models of protocellular membrane evolution with Jack Szostak (Lin*, Kamat* et. al. 2018).
Non-research Interests
I like spending time outside, and am always on the lookout for a good coffee or sandwich. I also enjoy reading/watching/looking at/listening to/discussing literature, films, art, and music.